-
Notifications
You must be signed in to change notification settings - Fork 5
ProgressReport
Shreyas edited this page Mar 31, 2014
·
17 revisions
The following is an update on the progress of Obidroid in terms of development
- Looking at the cluster analysis, the question is:
- what is special about the apps in cluster 1 and 7?
- You can look at those and then do something else with the other apps the are lumped together in 2-6. This might be worth digging into more.
- Here is a way to better assess your classification algorithm. Split it into 50/50 positive and negative tests.
- Based on the clustering results, choose some of the harder to discriminate items to be the positive items you compare against. Then see how well the classification algorithm works on the 50-50 split."
- We proceeded to dig deeper into each feature taken 1 at a time (Univariate Analysis) and taken 2 at a time (Bivariate Analysis)
-
Univariate Analysis
- We present histograms of features grouped on
fairandunfairapp Labels
- We present histograms of features grouped on
-
Bivariate Analysis
- Here we plotted plots for all combinations of features taken 2 at a time
- 2 kinds of plots are presented :
- hexbin plots
-
density plots (based on
kde)
- Also instead of scatter plot, we chose to use hexbin plots where the dots are binned as hexagon points.
- This was because we saw in our scatter plots a lot of points overlapped. And hence the plot was a little deceiving.
- We could have either added
jitterto the scatter plot to see all the points, but that felt like corrupting the data - We chose to bin the points with an
alphavalue in color shade. And hence a darker point represents that there are multiple points in those positions.
- We could have either added
- This was because we saw in our scatter plots a lot of points overlapped. And hence the plot was a little deceiving.
-
Univariate Analysis
- Did unsupervised learning (
clustering) on the features to understand how they were interacting.- We had already done k-means clustering previously.
- We proceeded to do MDS clustering in an aim to find how many apps were similar and dissimilar based on our feature extraction.
- Although we succeeded to plot the apps using
matplotlib, it was harder to discern the app names after the plot. - Hence we proceeded to develop an interactive
D3jsbased plot, which- which can be zoomed in
-
mouseoverinteraction gives App Names as tooltip. - This plotting method is an engine, and can be used for various other plots like
PCA, whose code is implemented. But we present here only the MDS output pertaining to our inquiry.
- Although we succeeded to plot the apps using
- We also plotted 2-dimensional Dendrogram, but frankly as with most dendrograms, it is harder to draw conclusions.
- Did supervised learning to find the most suitable classifier algorithms.
- The code takes 2 approaches
-
Equal Split Approach
- Divides the labeled apps into
fairandunfair - Splits
fairapps into multiple parts with the size ofunfairapps - All features are scaled using MinMaxScaler
- Randomly shuffle sample. Overall size is
46 - Trains on first
36apps and tests on last10. - Calculates classifier outputs for each classifier on each split
- No cross validation is performed.
- The results are tabulated in table below.
- To make the results comparable, the splits are performed first and each classifier is applied to the same split
- Divides the labeled apps into
-
All Apps
- We also apply all the same classifiers to the overall dataset.
- All features are scaled using MinMaxScaler
- Sample dataset is randomly shuffled.
-
k=4fold CrossValidation is performed - Classifier performance for each classifier in each fold is tabulated below.
- Since the folds are performed for each classifier separately, after random shuffling, performance in each fold for separate classifiers may or may not be comparable
-
Equal Split Approach
- Average precision for each classification operation is done below.
-
Adjusted average is calculated, using the below mentioned philosophy:
- As we have maintained from the start, false positives aren't an issue with us, but false negatives are.
- So we calculated adjusted average by
(TP + TN + FP)/Total- Where
TP=True Positive,TN=True Negative,FP = False Positive.
- Where
- The code takes 2 approaches
-
Univariate Analysis
- This was done after talking to the FTC as this is the approach they currently take while sifting through the app store. They search for all apps based on 1 particular attribute.
- Analyze each app on 1 particular feature, although it should be noted that we have added more features of our own extracted from app attributes.
- We were mostly looking for a feature that really stood out for
unfairapps.- There is no conclusive feature generally on their own that stand out for
unfairapps.
- There is no conclusive feature generally on their own that stand out for
-
Bivariate Analysis
- We found strong correlations between:
-
adjectiveCountxcountCapital -
revLengthxcountCapital
-
- and loose correlations between:
-
adjectiveCountxrevLength
-
- To develop a more parsimonious statistical model, we might want to combine these features into their product and drop these individual features.
- But for now, our classifiers are based on these individual features.
- We found strong correlations between:
-
Clustering
- Based on the MDS plots:
- there are some apps that are really very different from others and hence are easier to spot
- there are some apps that are very similar to other fair apps. This may not necessarily be a bad thing
- An example is
Whatsapp, which is in fact a very popular app but looking at the reviews we labeled it asunfair, which in this case is more likely to be afalse positive.
- An example is
- Based on the MDS plots:
-
Classifier
- In the equal split exploration
-
Gaussian Naive Bayesand(k=3)NearestNeighborswere the best bet in most of the splits.- Especially
GNBhad significantly lowerfalse negatives
- Especially
-
SVM-linear&SVM-NonLinear(rbf kernel) did OK mostly but failed quite spectacularly in others- It could also be due to very small size of the training data.
-
- In overall exploration
-
SVM-linearwas the most spectacular with the least false negatives -
RandomForestwas also a lot better than most.
-
- For our features there is not a lot of difference between
kNN uniformorkNN distanceweighted algorithms.
- In the equal split exploration
- The data was prepared from previously extracted
exports/appFeatures.csvusing our crawler and scraper scripts. - The data was then casted to appropriate datatypes explicitly to avoid any mistakes
- All variables in the data (i.e. the features) were then normalized using
min-maxnormalization.
For detailed feature descriptions, please refer Obidroid Project Report
Following is an exploration of each variable taken one-by-one at a time.
Here we explore the distributions of each data/feature
| Feature | Description | Hist (fair/unfair) |
|---|---|---|
adjectiveCount |
count of all adjectives | 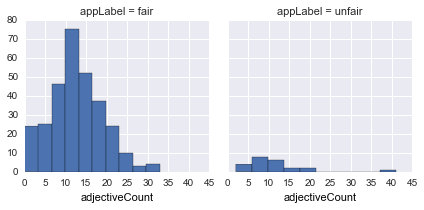 |
hasPrivacy |
Does it have a valid privacy url | 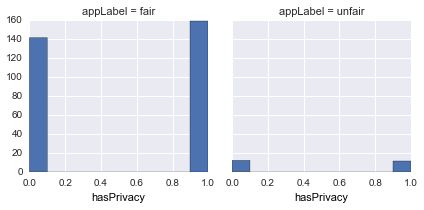 |
revLength |
Total characters in a review | 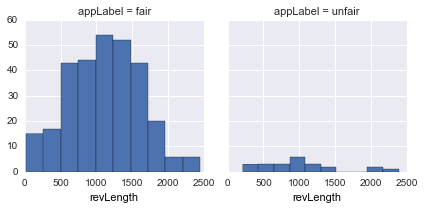 |
installs |
Total installs of an app | 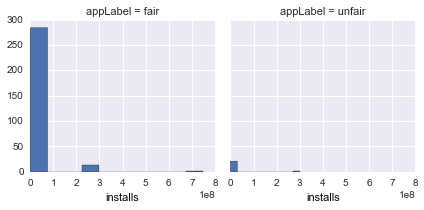 |
revSent |
Aggregate review sentiment | 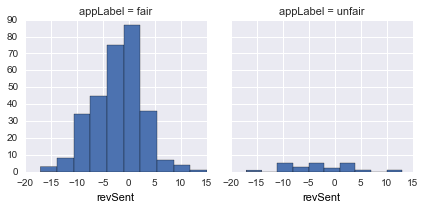 |
countCapital |
Count capital characters in a sentence | 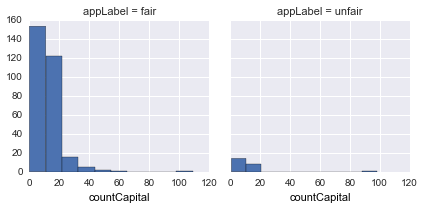 |
hasDeveloperWebsite |
Developer website |  |
hasDeveloperEmail |
Developer email |  |
avgRating |
average app Rating | 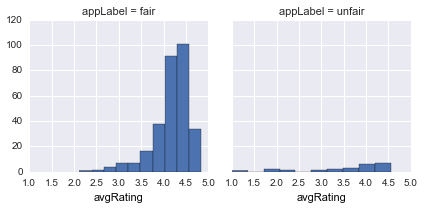 |
- It is hard to draw an inference about
fair/unfairbut just looking at 1 feature alone. -
adjectiveCounttapers in bothfairandunfairfor large number of adjectives, so with higher adjectives it might seem it is more likely to be unfair -
hasPrivacyis almost evenly distributed for bothfair/unfair -
revLengthseems to be uniform inunfairapps -
revSentforfair/unfairdistribution is quite similar -
avgRatingis generally skewed towards higher values for bothfair/unfair
| Feature1 x Feature 2 | Correlation Plot (as hexagon bins) | Density Plots (KDE) |
|---|---|---|
adjectiveCount x hasPrivacy
|
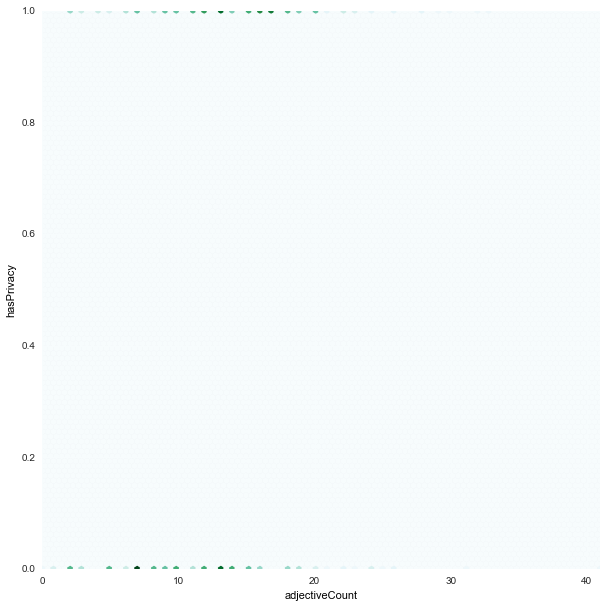 |
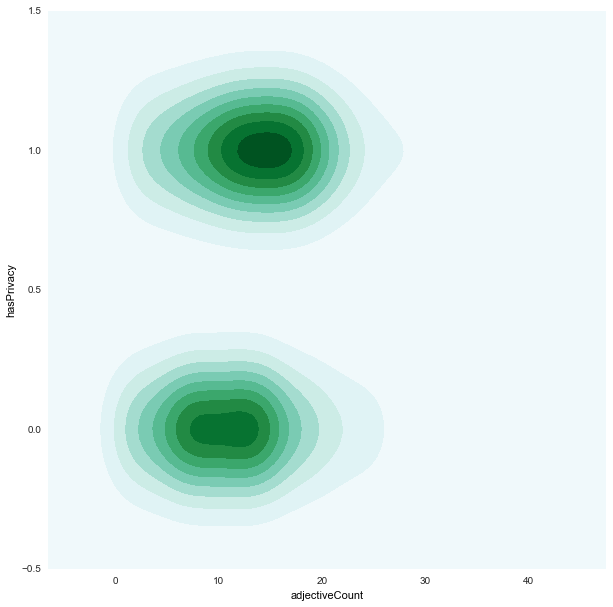 |
adjectiveCount x countCapital
|
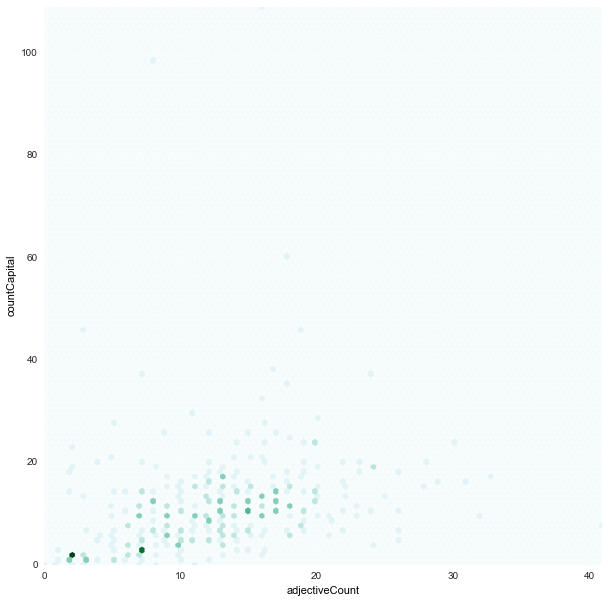 |
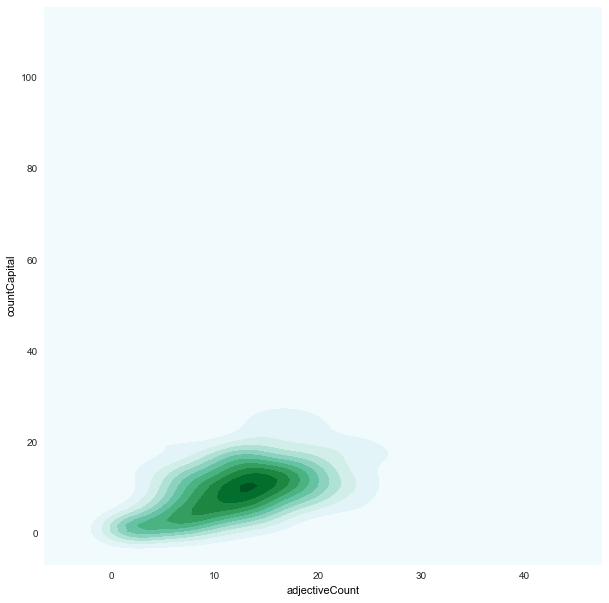 |
adjectiveCount x hasDeveloperEmail
|
 |
no plot [^note-2] |
adjectiveCount x hasDeveloperWebsite
|
 |
no plot [^note-2] |
adjectiveCount x hasPrivacy
|
 |
 |
adjectiveCount x installs
|
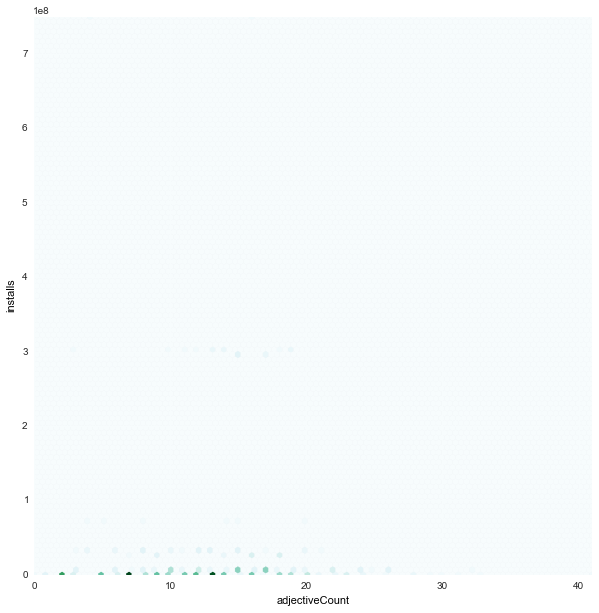 |
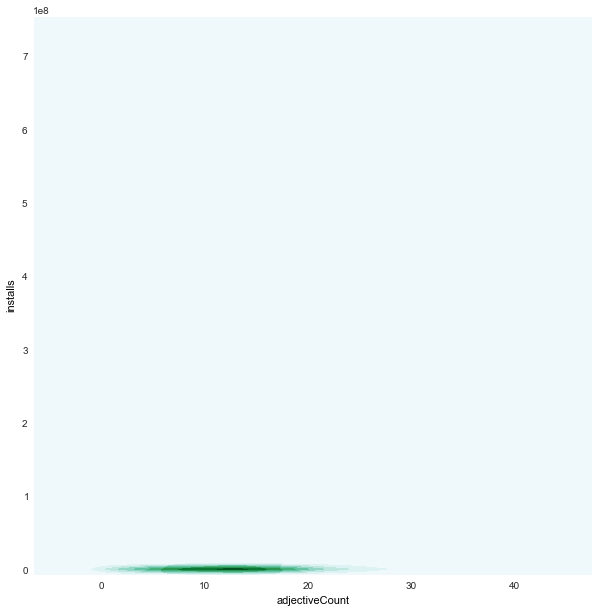 |
adjectiveCount x revLength
|
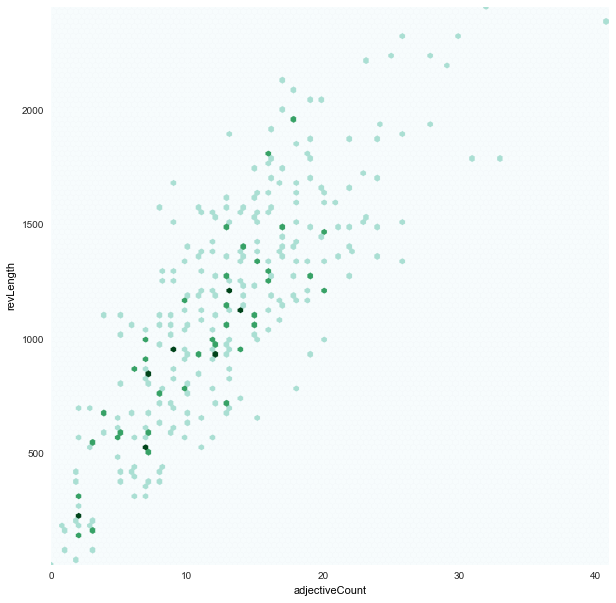 |
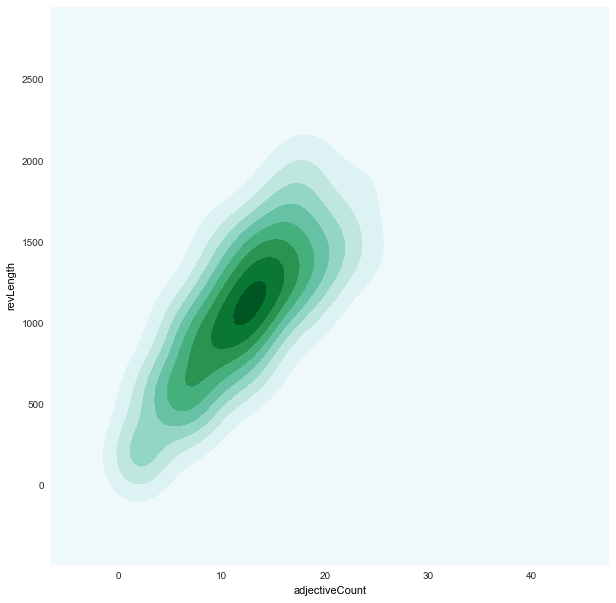 |
adjectiveCount x revSent
|
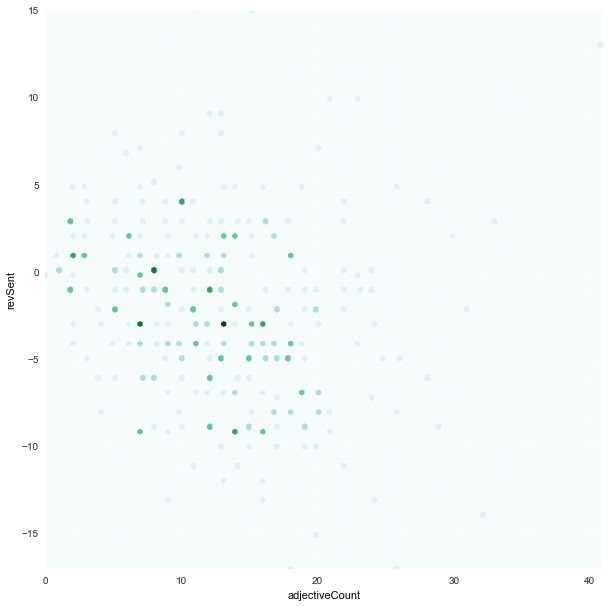 |
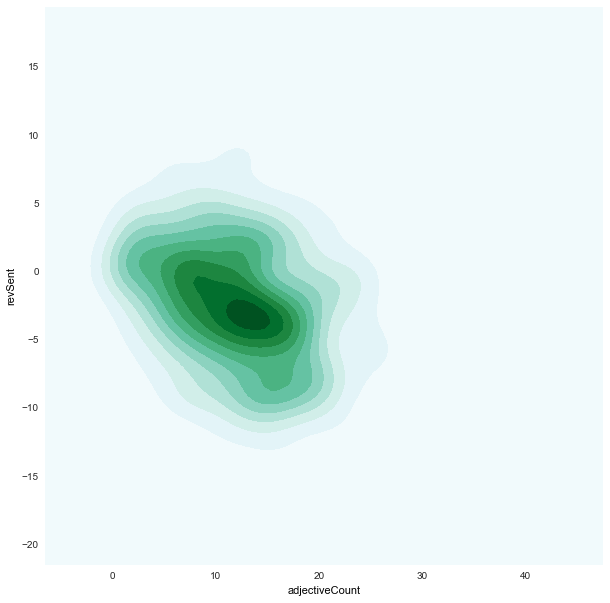 |
countCapital x avgRating
|
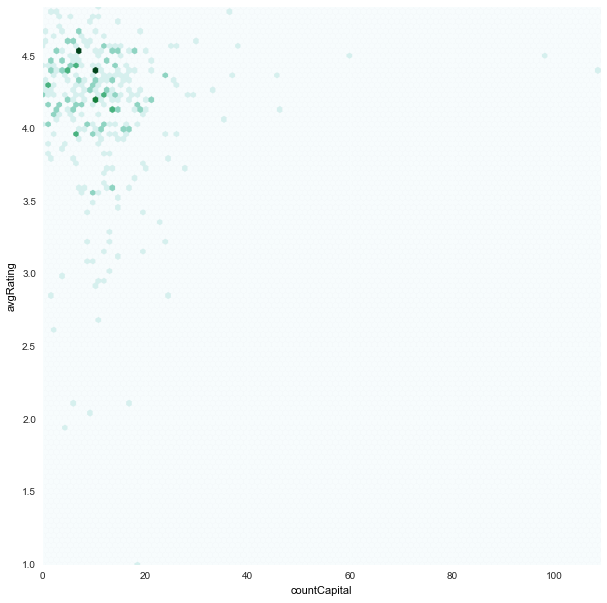 |
 |
countCapital x hasDeveloperEmail
|
 |
no plot [^note-2] |
countCapital x hasDeveloperWebsite
|
 |
no plot [^note-2] |
hasDeveloperEmail x avgRating
|
 |
no plot [^note-2] |
hasDeveloperWebsite x avgRating
|
 |
no plot [^note-2] |
hasDeveloperWebsite x hasDeveloperEmail
|
no plot [^note-2] |
no plot [^note-2] |
hasPrivacy x avgRating
|
 |
 |
hasPrivacy x countCapital
|
 |
 |
hasPrivacy x hasDeveloperEmail
|
 |
no plot [^note-2] |
hasPrivacy x hasDeveloperWebsite
|
 |
no plot [^note-2] |
hasPrivacy x installs
|
 |
no plot [^note-2] |
hasPrivacy x revLength
|
 |
no plot [^note-2] |
hasPrivacy x revSent
|
 |
 |
installs x avgRating
|
 |
 |
installs x countCapital
|
 |
 |
installs x countCapital
|
 |
 |
installs x hasDeveloperEmail
|
 |
no plot [^note-2] |
installs x hasDeveloperWebsite
|
 |
no plot [^note-2] |
revLength x avgRating
|
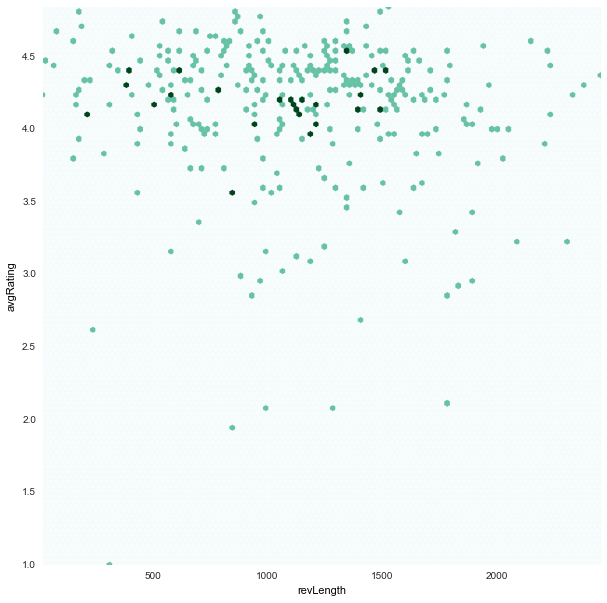 |
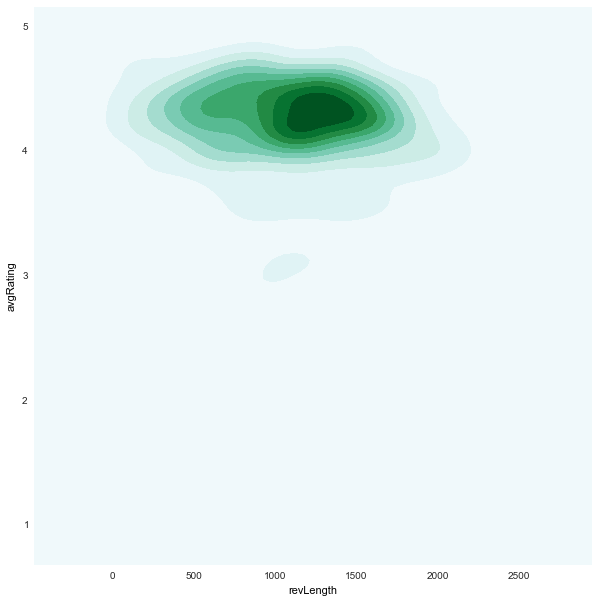 |
revLength x countCapital
|
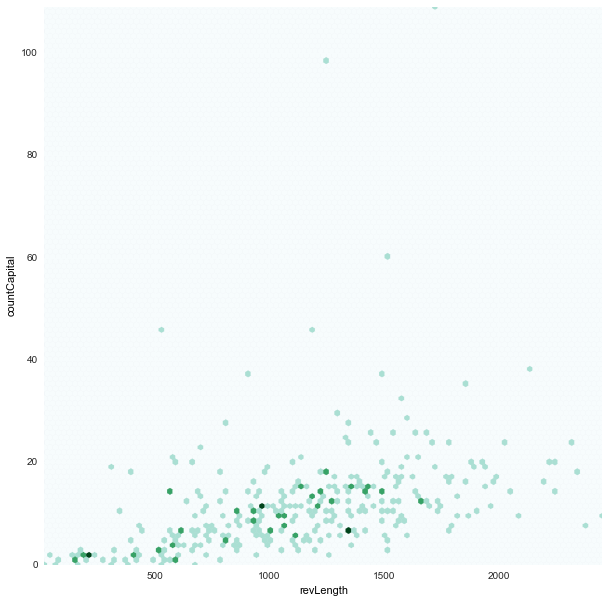 |
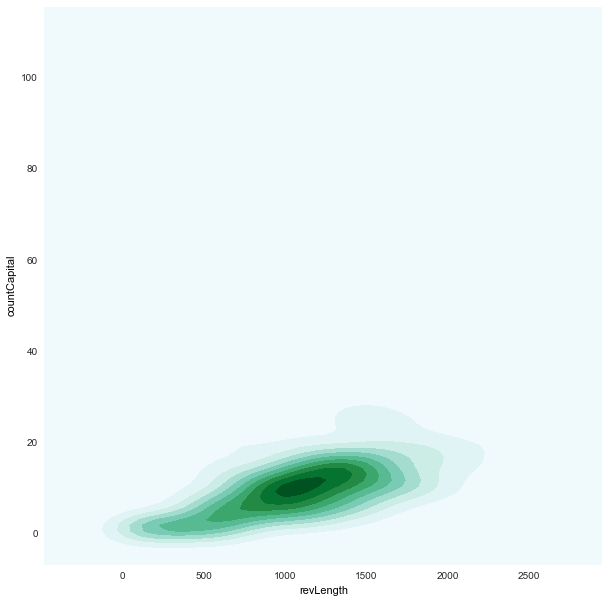 |
revLength x hasDeveloperEmail
|
 |
no plot [^note-2] |
revLength x hasDeveloperWebsite
|
 |
no plot [^note-2] |
revLength x revSent
|
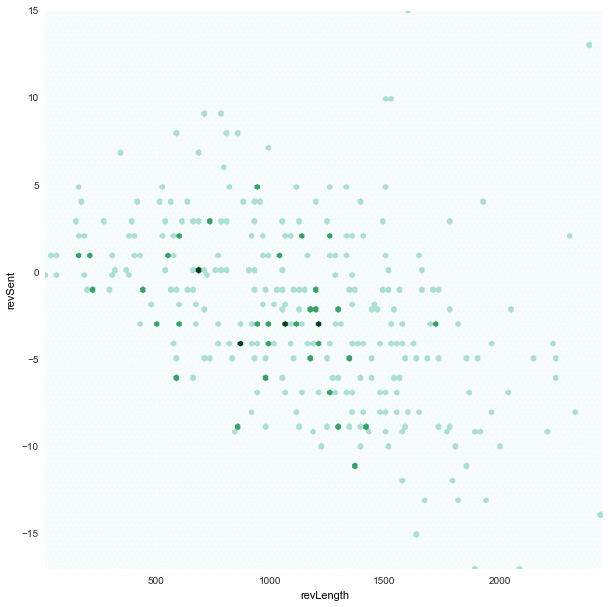 |
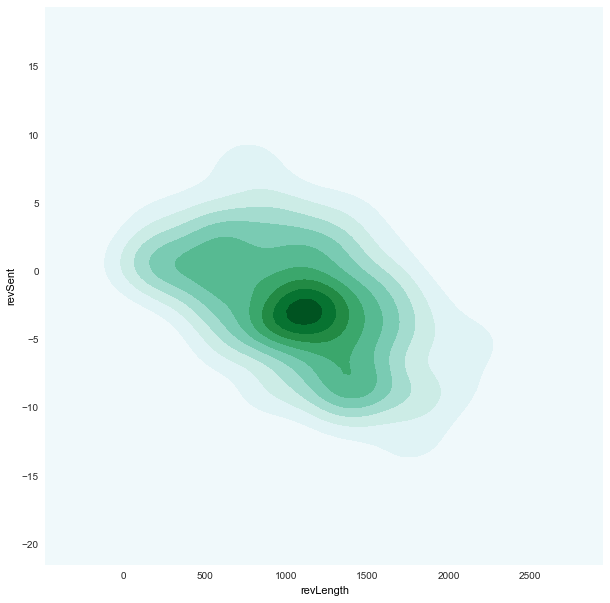 |
revSent x avgRating
|
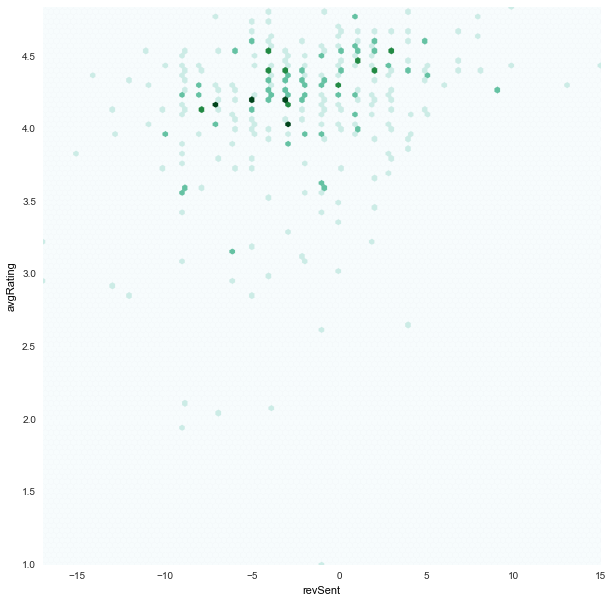 |
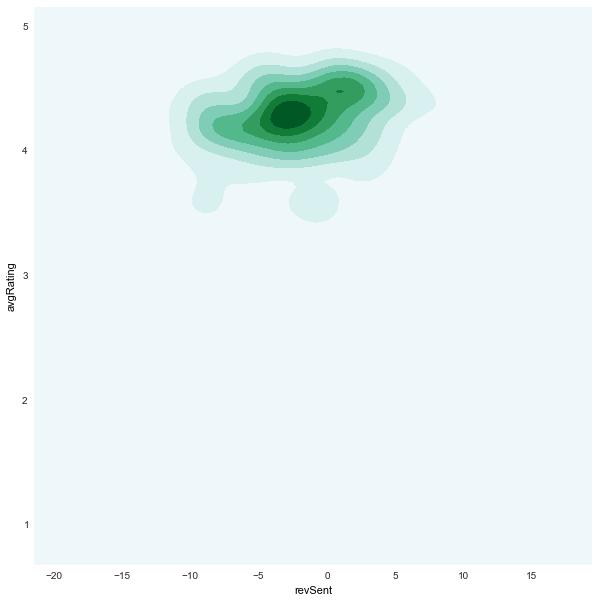 |
revSent x countCapital
|
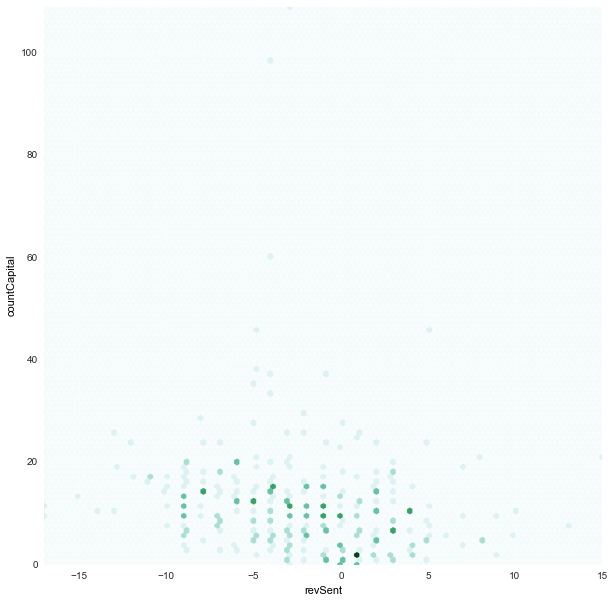 |
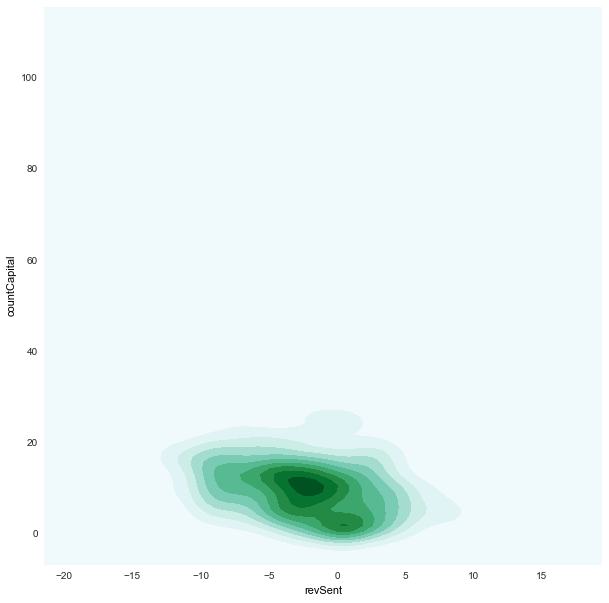 |
revSent x hasDeveloperEmail
|
 |
no plot [^note-2] |
revSent x hasDeveloperWebsite
|
 |
no plot [^note-2] |
Static Plot:
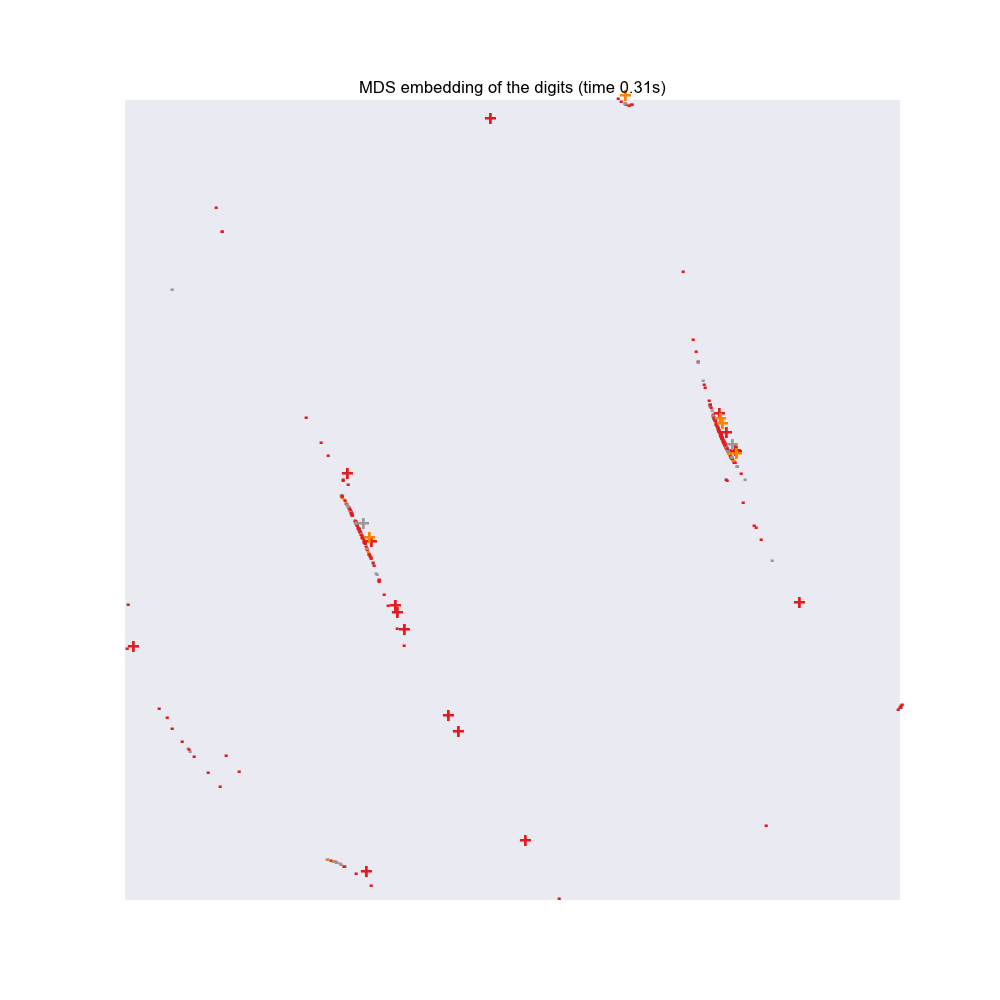
-
+representunfairapp -
.representfairapp
- refer plot at bottom.
- use trackpad to zoom
- use mouseover to view app titles.
Dendrogram static plot
Don't know how to interpret.
All other plots can be viewed at Github repo.
Our process:
- split the entire app sample into
fairapps (300) andunfairapps (23) - split the fair apps sample into splits of the size of the
unfairapps.- Total 13 splits
- Combined each fair app split with the unfair apps, to make the sample set for classification
- randomly shuffled the classification sample
- trained on
n_sample=36apps and tested ontotal-n_sample = 10apps for each split - Calculated classifier reports for each split
- Explicit SplitClassifierReport
| Split # | Algorithm | Avg Precision | Avg Accuracy (adjusted) | Confusion Matrix [[TP FN][FP TN]]
|
|---|---|---|---|---|
0th |
kNN (wt = distance)
|
0.50 |
0.70 |
[[2 3] [2 3]] |
0th |
kNN (wt = uniform)
|
0.50 |
0.70 |
[[2 3] [2 3]] |
0th |
GaussianNB | 0.60 |
0.80 |
[[3 2][2 3]] |
0th |
DecisionTreeClassifier | 0.50 |
0.80 |
[[3 2][3 2]] |
0th |
RandomForest ( AdaBoostClassifier) |
0.60 |
0.80 |
[[3 2][2 3]] |
0th |
SVM-linear (SVC) |
0.62 |
0.70 |
[[2 3][1 4]] |
0th |
SVM-Nonlinear (NuSVC) |
0.60 |
0.80 |
[[3 2][2 3]] |
1st |
kNN (wt = distance)
|
0.75 |
0.80 |
[[6 2][1 1]] |
1st |
kNN (wt = uniform)
|
0.80 |
0.90 |
[[7 1][1 1]] |
1st |
GaussianNB | 0.64 |
1.0 |
[[8 0][2 0]] |
1st |
DecisionTreeClassifier | 0.53 |
0.60 |
[[4 4][2 0]] |
1st |
RandomForest ( AdaBoostClassifier) |
0.75 |
0.80 |
[[6 2][1 1]] |
1st |
SVM-linear (SVC) |
0.04 |
0.2 |
[[0 8][0 2]] |
1st |
SVM-Nonlinear (NuSVC) |
0.80 |
0.90 |
[[7 1][1 1]] |
2nd |
kNN (wt = distance)
|
0.71 |
0.90 |
[[4 1][2 3]] |
2nd |
kNN (wt = uniform)
|
0.71 |
0.90 |
[[4 1][2 3]] |
2nd |
GaussianNB | 0.38 |
0.8 |
[[3 2][4 1]] |
2nd |
DecisionTreeClassifier | 0.29 |
0.70 |
[[2 3][4 1]] |
2nd |
RandomForest ( AdaBoostClassifier) |
0.29 |
0.70 |
[[2 3][4 1]] |
2nd |
SVM-linear (SVC) |
0.50 |
0.9 |
[[1 4][1 4]] |
2nd |
SVM-Nonlinear (NuSVC) |
0.60 |
0.80 |
[[3 2][2 3]] |
3rd |
kNN (wt = distance)
|
0.80 |
0.90 |
[[4 1][1 4]] |
3rd |
kNN (wt = uniform)
|
0.80 |
0.90 |
[[4 1][1 4]] |
3rd |
GaussianNB | 0.81 |
1.0 |
[[5 0][3 2]] |
3rd |
DecisionTreeClassifier | 0.60 |
0.80 |
[[3 2][2 3]] |
3rd |
RandomForest ( AdaBoostClassifier) |
0.40 |
0.70 |
[[2 3][3 2]] |
3rd |
SVM-linear (SVC) |
0.81 |
0.7 |
[[2 3][0 5]] |
3rd |
SVM-Nonlinear (NuSVC) |
0.71 |
0.80 |
[[3 2][1 4]] |
4th |
kNN (wt = distance)
|
0.68 |
0.70 |
[[4 3][1 2]] |
4th |
kNN (wt = uniform)
|
0.68 |
0.70 |
[[4 3][1 2]] |
4th |
GaussianNB | 0.84 |
1.0 |
[[7 0][2 1]] |
4th |
DecisionTreeClassifier | 0.62 |
0.60 |
[[3 4][1 2]] |
4th |
RandomForest ( AdaBoostClassifier) |
0.68 |
0.70 |
[[4 3][1 2]] |
4th |
SVM-linear (SVC) |
0.07 |
0.30 |
[[0 7][1 2]] |
4th |
SVM-Nonlinear (NuSVC) |
0.55 |
0.50 |
[[2 5][1 2]] |
5th |
kNN (wt = distance)
|
0.62 |
0.70 |
[[4 1][3 2]] |
5th |
kNN (wt = uniform)
|
0.62 |
0.70 |
[[4 1][3 2]] |
5th |
GaussianNB | 0.25 |
1.0 |
[[5 0][5 0]] |
5th |
DecisionTreeClassifier | 0.71 |
0.80 |
[[3 2][1 4]] |
5th |
RandomForest ( AdaBoostClassifier) |
0.71 |
0.80 |
[[3 2][1 4]] |
5th |
SVM-linear (SVC) |
0.60 |
0.80 |
[[3 2][2 3]] |
5th |
SVM-Nonlinear (NuSVC) |
0.71 |
0.90 |
[[4 1][2 3]] |
6th |
kNN (wt = distance)
|
0.80 |
0.90 |
[[5 1][1 3]] |
6th |
kNN (wt = uniform)
|
0.80 |
0.90 |
[[5 1][1 3]] |
6th |
GaussianNB | 0.80 |
1.0 |
[[6 0][3 1]] |
6th |
DecisionTreeClassifier | 0.57 |
0.60 |
[[2 4][1 3]] |
6th |
RandomForest ( AdaBoostClassifier) |
0.71 |
0.80 |
[[3 2][1 4]] |
6th |
SVM-linear (SVC) |
0.16 |
0.40 |
[[0 6][0 4]] |
6th |
SVM-Nonlinear (NuSVC) |
0.85 |
1.0 |
[[6 0][2 2]] |
7th |
kNN (wt = distance)
|
0.80 |
1.0 |
[[4 0][4 2]] |
7th |
kNN (wt = uniform)
|
0.83 |
1.0 |
[[4 0][3 3]] |
7th |
GaussianNB | 0.16 |
1.0 |
[[4 0][6 0]] |
7th |
DecisionTreeClassifier | 0.52 |
0.80 |
[[2 2][3 3]] |
7th |
RandomForest ( AdaBoostClassifier) |
0.45 |
0.90 |
[[3 1][5 1]] |
7th |
SVM-linear (SVC) |
0.16 |
1.0 |
[[4 0][6 0]] |
7th |
SVM-Nonlinear (NuSVC) |
0.45 |
0.90 |
[[3 1][5 1]] |
8th |
kNN (wt = distance)
|
0.63 |
0.90 |
[[1 1][5 3]] |
8th |
kNN (wt = uniform)
|
0.63 |
0.90 |
[[1 1][5 3]] |
8th |
GaussianNB | 0.63 |
0.90 |
[[1 1][5 3]] |
8th |
DecisionTreeClassifier | 0.42 |
0.90 |
[[1 1][7 1]] |
8th |
RandomForest ( AdaBoostClassifier) |
0.56 |
0.90 |
[[1 1][6 2]] |
8th |
SVM-linear (SVC) |
0.04 |
1.0 |
[[2 0][8 0]] |
8th |
SVM-Nonlinear (NuSVC) |
0.56 |
0.90 |
[[1 1][6 2]] |
9th |
kNN (wt = distance)
|
0.71 |
0.90 |
[[4 1][2 3]] |
9th |
kNN (wt = uniform)
|
0.71 |
0.90 |
[[4 1][2 3]] |
9th |
GaussianNB | 0.78 |
1.0 |
[[5 0][4 1]] |
9th |
DecisionTreeClassifier | 0.22 |
0.90 |
[[4 1][5 0]] |
9th |
RandomForest ( AdaBoostClassifier) |
0.25 |
1.0 |
[[5 0][5 0]] |
9th |
SVM-linear (SVC) |
0.25 |
1.0 |
[[5 0][5 0]] |
9th |
SVM-Nonlinear (NuSVC) |
0.78 |
1.0 |
[[5 0][4 1]] |
10th |
kNN (wt = distance)
|
0.92 |
0.60 |
[[5 4][0 1]] |
10th |
kNN (wt = uniform)
|
0.93 |
0.7 |
[[6 3][0 1]] |
10th |
GaussianNB | 0.93 |
0.80 |
[[7 2][0 1]] |
10th |
DecisionTreeClassifier | 0.91 |
0.30 |
[[2 7][0 1]] |
10th |
RandomForest ( AdaBoostClassifier) |
0.91 |
0.2 |
[[1 8][0 1]] |
10th |
SVM-linear (SVC) |
0.01 |
0.1 |
[[0 9][0 1]] |
10th |
SVM-Nonlinear (NuSVC) |
0.92 |
0.5 |
[[4 5][0 1]] |
11th |
kNN (wt = distance)
|
0.43 |
0.80 |
[[2 2][4 2]] |
11th |
kNN (wt = uniform)
|
0.78 |
1.0 |
[[4 0][5 1]] |
11th |
GaussianNB | 0.45 |
0.90 |
[[3 1][5 1]] |
11th |
DecisionTreeClassifier | 0.31 |
0.80 |
[[2 2][5 1]] |
11th |
RandomForest ( AdaBoostClassifier) |
0.31 |
0.80 |
[[2 2][5 1]] |
11th |
SVM-linear (SVC) |
0.16 |
1.0 |
[[4 0][6 0]] |
11th |
SVM-Nonlinear (NuSVC) |
0.45 |
0.90 |
[[3 1][5 1]] |
12th |
kNN (wt = distance)
|
0.60 |
0.80 |
[[4 2][2 2]] |
12th |
kNN (wt = uniform)
|
0.60 |
0.80 |
[[4 2][2 2]] |
12th |
GaussianNB | 0.33 |
0.90 |
[[5 1][4 0]] |
12th |
DecisionTreeClassifier | 0.57 |
0.90 |
[[5 1][3 1]] |
12th |
RandomForest ( AdaBoostClassifier) |
0.80 |
1.0 |
[[6 0][3 1]] |
12th |
SVM-linear (SVC) |
0.16 |
0.4 |
[[0 6][0 4]] |
12th |
SVM-Nonlinear (NuSVC) |
0.30 |
0.80 |
[[4 2][4 0]] |
- scaled the features on
MinMaxScaler - performed
k=4fold Cross-Validation - Used same classifiers
- Explicit OverallClassifierReport
| Fold # | Algorithm | Avg Precision | Avg Accuracy (adjusted) | Confusion Matrix [[TP FN][FP TN]]
|
|---|---|---|---|---|
1st |
kNN (wt = distance)
|
0.97 |
0.95 |
[[74 4][ 1 1]] |
2nd |
kNN (wt = distance)
|
0.93 |
0.9875 |
[[76 1][ 3 0]] |
3rd |
kNN (wt = distance)
|
0.90 |
0.9875 |
[[75 1][ 4 0]] |
4th |
kNN (wt = distance)
|
0.79 |
0.9875 |
[[66 1][12 1]] |
1st |
kNN (wt = uniform)
|
0.97 |
0.975 |
[[76 2][ 1 1]] |
2nd |
kNN (wt = uniform)
|
0.93 |
1.00 |
[[77 0][ 3 0]] |
3rd |
kNN (wt = uniform)
|
0.90 |
1.00 |
[[76 0][ 4 0]] |
4th |
kNN (wt = uniform)
|
0.79 |
0.9875 |
[[66 1][12 1]] |
1st |
GaussianNB | 0.96 |
0.9125 |
[[71 7][ 1 1]] |
2nd |
GaussianNB | 0.96 |
0.95 |
[[73 4][ 1 2]] |
3rd |
GaussianNB | 0.90 |
1.00 |
[[76 0][ 4 0]] |
4th |
GaussianNB | 0.64 |
0.5875 |
[[14 53][ 5 8]] |
1st |
DecisionTreeClassifier | 0.95 |
0.875 |
[[68 10][ 2 0]] |
2nd |
DecisionTreeClassifier | 0.92 |
0.9125 |
[[70 7][ 3 0]] |
3rd |
DecisionTreeClassifier | 0.93 |
0.8625 |
[[65 11][ 2 2]] |
4th |
DecisionTreeClassifier | 0.82 |
0.975 |
[[65 2][10 3]] |
1st |
RandomForest ( AdaBoostClassifier) |
0.95 |
0.9625 |
[[75 3][ 2 0]] |
2nd |
RandomForest ( AdaBoostClassifier) |
0.93 |
0.9875 |
[[76 1][ 3 0]] |
3rd |
RandomForest ( AdaBoostClassifier) |
0.90 |
1.0 |
[[76 0][ 4 0]] |
4th |
RandomForest ( AdaBoostClassifier) |
0.82 |
0.975 |
[[65 2][10 3]] |
1st |
SVM-linear (SVC) |
0.95 |
1.0 |
[[78 0][ 2 0]] |
2nd |
SVM-linear (SVC) |
0.93 |
1.0 |
[[77 0][ 3 0]] |
3rd |
SVM-linear (SVC) |
0.90 |
1.0 |
[[76 0][ 4 0]] |
4th |
SVM-linear (SVC) |
0.70 |
1.0 |
[[67 0][13 0]] |
1st |
SVM-Nonlinear (NuSVC) [^note-1] |
- |
- |
- |
2nd |
SVM-Nonlinear (NuSVC) [^note-1] |
- |
- |
- |
3rd |
SVM-Nonlinear (NuSVC) [^note-1] |
- |
- |
- |
4th |
SVM-Nonlinear (NuSVC) [^note-1] |
- |
- |
- |
| Classifier Name | Classifier Parameters |
|---|---|
kNN (wt = distance)
|
(algorithm=auto, leaf_size=30, metric=minkowski, n_neighbors=3, p=2, weights=distance) |
kNN (wt = uniform)
|
(algorithm=auto, leaf_size=30, metric=minkowski, n_neighbors=3, p=2, weights=uniform) |
| GaussianNB | default |
| DecisionTreeClassifier | (compute_importances=None, criterion=gini, max_depth=None, max_features=None, min_density=None, min_samples_leaf=1, min_samples_split=2, random_state=None, splitter=best) |
RandomForest ( AdaBoostClassifier) |
AdaBoostClassifier(algorithm=SAMME, base_estimator=DecisionTreeClassifier(compute_importances=None, criterion=gini, max_depth=1, max_features=None, min_density=None, min_samples_leaf=1, min_samples_split=2, random_state=None, splitter=best), base_estimator__compute_importances=None, base_estimator__criterion=gini, base_estimator__max_depth=1, base_estimator__max_features=None, base_estimator__min_density=None, base_estimator__min_samples_leaf=1, base_estimator__min_samples_split=2, base_estimator__random_state=None, base_estimator__splitter=best, learning_rate=1.0, n_estimators=200, random_state=None) |
SVM-linear (SVC) |
(C=1.0, cache_size=200, class_weight=None, coef0=0.0, degree=3, gamma=0.0, kernel=rbf, max_iter=-1, probability=False, random_state=None, shrinking=True, tol=0.001, verbose=False) |
SVM-Nonlinear (NuSVC) |
(cache_size=200, coef0=0.0, degree=3, gamma=0.0, kernel=rbf, max_iter=-1, nu=0.5, probability=False, random_state=None, shrinking=True, tol=0.001, verbose=False) |
- [^note-1]: Some bug in the Non-Linear SVM implementation for all apps needs to be resolved. Currently it exits with an error
ValueError: specified nu is infeasible - [^note-2]: For some boolean variables no density plots are generated using
kde, but for others it does. We don't really know why. Would love your comment on it. The general culprits seem to behasDeveloperEmailandhasDeveloperWebsite.hasPrivacyworks mostly fine.