PyTorch implementation of "SPD-CNN: A Plain CNN-Based Model Using the Symmetric Positive Definite Matrices for Cross-Subject EEG Classification with Meta-Transfer-Learning"
This repository contains the PyTorch implementation for the Paper "SPD-CNN: A Plain CNN-Based Model Using the Symmetric Positive Definite Matrices for Cross-Subject EEG Classification with Meta-Transfer-Learning"
This repository is mainly modified from the Github repository of "Meta-transfer-learning"(https://github.com/sabinechen/meta-transfer-learning). If you have any questions on this repository or the related paper, feel free to create an issue or send me an email ([email protected]).
To build a robust classification algorithm for a calibration-less BCI systemcalibration-less BCI system, we propose an end-to-end model that transforms the EEG signals into symmetric positive definite (SPD) matrices and captures the features of SPD matrices by using a CNN. To avoid the time-consuming calibration and ensure the application of the proposed model, we use the meta-transfer-learning (MTL) method to learn the essential features from different subjects.The main Figure are shown below.
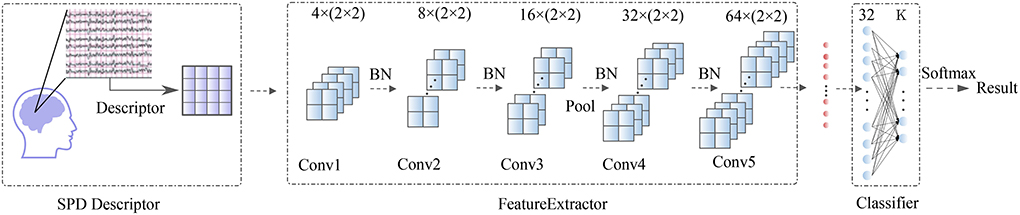
Figure1: Overall visualization of the SPD-CNN architecture. .
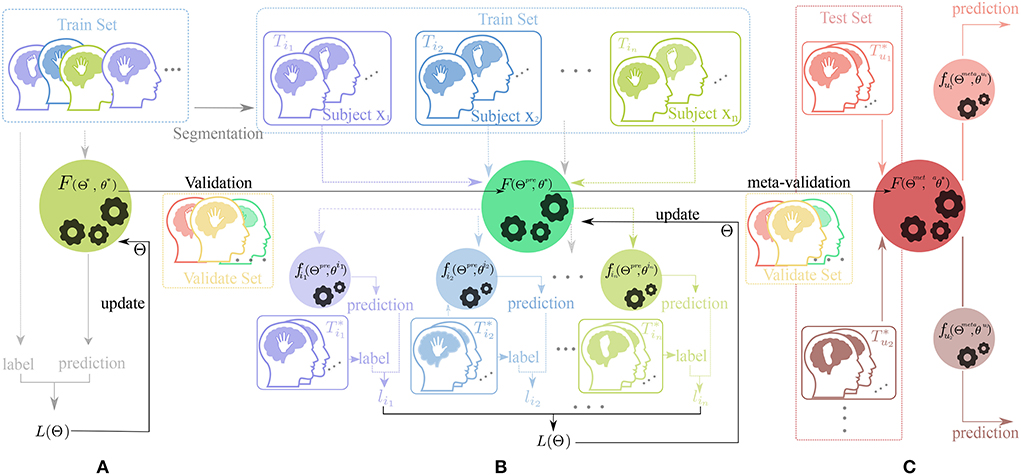
Figure2: Workflow of our training framework .
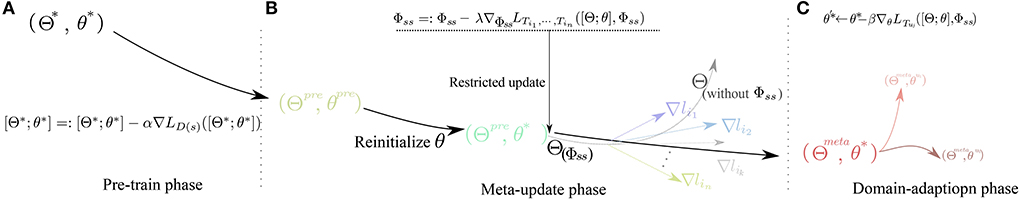
Figure3: Diagram of parameters variation through the learning process in different phases.
A whole visualization and full description of SPD-CNN model can be found in Figure 1.The Workflow of our training framework are shown in Figuer2 and Figuer 3.
In order to run this repository, we advise you to install python 3.7 and PyTorch 1.4.0 with Anaconda. You may download Anaconda and read the installation instruction on their official website: https://www.anaconda.com/download/
Create a new environment and install PyTorch and on it:
conda create --name mtl-pytorch python=3.7
conda activate mtl-pytorch
conda install pytorch=1.4.0 Install other requirements(for generating the dataset and processing the data):smile::smile::
pip install parse,pillow,scipy,moabbThen use the python file in the Data_generator to download and process the datasets:smile:.
The data that support the findings of this study are openly available in https://github.com/NeuroTechX/moabb.
Please cite our paper if it is helpful to your work:
@article{chenspd,
title={SPD-CNN: A Plain CNN-Based Model Using the Symmetric Positive Definite Matrices for Cross-Subject EEG Classification with Meta-Transfer-Learning},
author={Chen, Lezhi and Yu, Zhuliang and Yang, Jian},
journal={Frontiers in Neurorobotics},
pages={168},
publisher={Frontiers}
}Our implementations use the source code from the following repositories and users 😄: